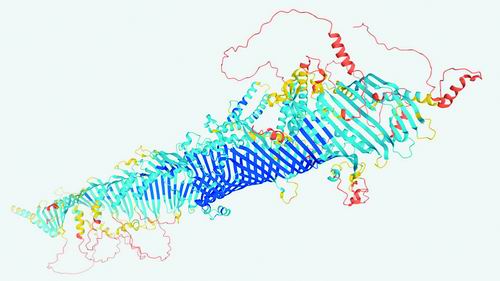
AI can now quickly and reliably predict the three-dimensional shape of most proteins.Image source: DEEPMIND
Recently, the University of Washington in the United States and DeepMind in the United Kingdom have respectively announced the results of years of work: advanced modeling programs that can predict the precise three-dimensional atomic structure of proteins and some molecular complexes. One of the research teams reported that they have used newly developed artificial intelligence (AI) programs to predict 350,000 protein structures from humans and 20 model organisms-such as E. coli, yeast, and fruit flies. In the next few months, they plan to include all the cataloged proteins on the model protein list, which has approximately 100 million molecules.
“This is quite amazing.” John Moult, a protein expert at the University of Maryland in the United States, said that he holds a competition called “Critical Protein Structure Prediction Method” (CASP) every two years. Moult said that for decades, structural biologists have dreamed that one day, computer models could increase the number of extremely precise protein shapes obtained from experimental methods such as X-ray crystallography. “I never thought this dream would come true.” Moult said.
This model called AlphaFold is the result of researchers at DeepMind, a British AI company affiliated with Google’s parent company Alphabet. In 2020, AlphaFold “swept” CASP. But DeepMind researchers did not disclose the theoretical details of drawing protein shapes, especially the underlying computer code of AlphaFold.
This situation has begun to change. On July 15, the Minkyung Baek and David Baker research groups of the University of Washington reported that they had created a highly accurate protein structure prediction program called RoseTTAFold and released it publicly. Related results are published online in “Science”. At the same time, “Nature” published a paper written by DeepMind researchers Demis Hassabis and John Jumper, announcing the details of AlphaFold.
Both programs use AI to identify folding patterns in a huge database of protein structures. These programs calculate the most likely structure of an unknown protein by considering the basic physical and biological rules of the interaction of adjacent amino acids in the protein. The paper shows that Baek and Baker used RoseTTAFold to create a structural database containing hundreds of G protein-coupled receptors (a common drug target).
And DeepMind researchers created 350,000 prediction structures, more than twice the results obtained by previous experimental methods. The researchers said that AlphaFold produced nearly 44% of the human protein structure, covering nearly 60% of the amino acids encoded by the human genome. AlphaFold determined that many other human proteins are “disordered,” which means that their shape is not a single structure.
In addition, DeepMind cooperated with the European Molecular Biology Laboratory to build a new protein prediction database, which can be accessed online for free. “It’s great to be able to provide this kind of service.” Baker said, “It really speeds up the pace of research.” Because the 3D structure of a protein largely determines its function, this database is convenient for biologists to clarify. How thousands of unknown proteins work. (Bunraku)
[
责编:武玥彤 ]
.